Abstract
Background
Despite recent drug development efforts, multi-drug resistance remains a major challenge in the treatment of AML. Understanding resistance mechanisms could help to design combinatorial therapies beneficial for AML patients. Earlier studies have shown that sensitivity of AML cells to the B-cell lymphoma-2 (BCL2) inhibitor venetoclax is associated with mutations in genes encoding chromatin modifiers, WT1 and IDH1/IDH2 (Konopleva et al, Cancer Disc. 2016). We have also observed that overexpression of a subset of HOXA and HOXB genes is linked to venetoclax sensitivity (Kontro et al, Leukemia 2016). Besides, induction of alternate cell survival mechanisms (e.g. BCLXL and MCL1), there is little knowledge of molecular indicators of venetoclax resistance. In this study, we used RNA sequence and ex vivo drug sensitivity data from a cohort of AML patients to identify gene expression profiles linked to venetoclax resistance.
Methods
RNA sequencing was performed on bone marrow or peripheral blood mononuclear cells (MNCs) collected from 112 AML patients and 4 healthy individuals. Sensitivity of MNCs from 18 AML patient samples was assessed to venetoclax by measuring cell viability after 72 h incubation with 6 different concentrations (1-1000 nM) using the CellTiter-Glo (CTG) assay. Free cytosolic calcium was measured in venetoclax-resistant (OCI-AML3, SHI-1) and sensitive cell lines (MOLM-13, Kasumi-1, all from DSMZ) using a calcium assay kit (BD Biosciences) after exposure to venetoclax (100 nM) for 2, 4 and 6 h. Statistical dependence between two variables was assessed by Pearson's correlation coefficient modeling.
Results
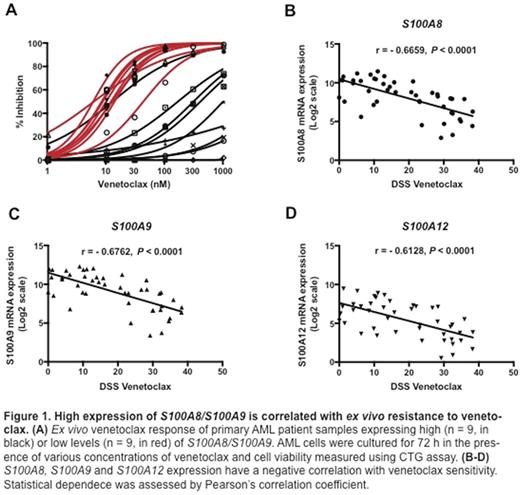
The S100 family is comprised of 21 different low molecular weight, calcium-binding proteins. Interestingly, nearly half of the S100 family members had elevated expression in AML patient samples as compared with healthy individuals, with S100A8 and S100A9 being the most highly expressed genes. Based on this observation and a previous analysis of overexpressed genes in venetoclax resistant samples (Kontro et al, Leukemia 2016), we studied correlation of S100 gene expression with resistance to venetoclax. Wetested sensitivity of AML patient samples with the highest versus lowest expression of the genes (n = 9, each group) to venetoclax and confirmed correlation of high gene expression with reduced drug sensitivity (Figure 1A). High expression of S100A8, S100A9 and S100A12 negatively correlated with sensitivity to venetoclax (Figure 1B-D). Over-expression of S100A8 and S100A9 on venetoclax-resistant cell lines (OCI-AML3, SHI-1) was also associated with failure to release free-cytosolic Ca2+ compared with venetoclax-sensitive cell lines (MOLM-13, Kasumi-1). The S100A8 and S100A9 proteins form a complex and bind free Ca2+, which can inhibit mitochondrial membrane depololarization and the initiation of apoptosis. Over expression of these genes in AML could therefore potentially contribute to the development of venetoclax resistance.
Conclusions
BCL2 inhibitors such as venetoclax are effective novel therapies for several malignancies, including AML. However, identifying responding patients and understanding mechanisms of resistance are important. Our results showing high expression of the S100A8 and S100A9 calcium-binding proteins correlates with AML cell resistance to venetoclax warrant further validation in a clinical setting. However, these data provide evidence of a novel mechanism of resistance to this class of drug.
Acknowledgment
RK and ML equal contribution.
Porkka: Celgene: Honoraria, Research Funding; Bristol-Myers Squibb: Honoraria, Research Funding; Novartis: Honoraria, Research Funding; Pfizer: Honoraria, Research Funding. Heckman: Celgene: Research Funding; Novartis: Research Funding; Orion Pharma: Research Funding; Pfizer: Research Funding; IMI2 project HARMONY: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal